Performance of classifiers¶
Classifier for HasNS trained on \(m_2 \leq 3 M_{\odot}\) to have an NS. 2H EoS used for remnant matter computation.
[1]:
import pickle
import pkg_resources
import numpy as np
import matplotlib.pyplot as plt
from astropy.table import Table
from ligo import em_bright
[2]:
with pkg_resources.resource_stream('ligo.data', 'knn_ns_classifier.pkl') as f:
clf_ns = pickle.load(f)
with pkg_resources.resource_stream('ligo.data', 'knn_em_classifier.pkl') as f:
clf_em = pickle.load(f)
[3]:
# sweep masses
mass1 = np.linspace(1, 100, 1000)
mass2 = np.linspace(1, 100, 1000)
t = Table(data=np.vstack((np.repeat(mass1, mass2.size),
np.tile(mass2, mass1.size))).T, names=('mass1', 'mass2'))
mask = t['mass1'] > t['mass2']
t = t[mask]
# vary spin, fix SNR
spins = Table(data=np.vstack((np.repeat(np.linspace(0, 1, 2), 2),
np.tile(np.linspace(0, 1, 2), 2))).T,
names=('chi1', 'chi2'))
[4]:
def make_plots(features, predictions, title, fig_idx):
fig_, idx = fig_idx
fig_.add_subplot(4, 1, idx)
# indices 0 and 1 correspond to mass1 and mass2 respectively
plt.scatter(features.T[0], features.T[1],
s=10, c=predictions)
plt.title(title)
plt.tight_layout()
plt.colorbar(label='Probability')
# plot chirp mass contours
m1 = np.linspace(1, 50, 100)
m2 = np.linspace(1, 50, 100)
M1, M2 = np.meshgrid(m1, m2)
Mc = (M1*M2)**(3./5.)/(M1 + M2)**(1./5.)
Mc = np.tril(Mc).T
CS = plt.contour(M1, M2, Mc, levels=[5, 6, 7, 8, 9],
colors='black', linewidths=1)
plt.clabel(CS, inline=True, fontsize=16)
plt.xlim((1, 50))
plt.ylim((1, 14))
plt.axhline(y=3.0, c='r')
plt.xlabel(r'$m_1$', fontsize=16)
plt.ylabel(r'$m_2$', fontsize=16)
plt.title(title)
plt.tight_layout()
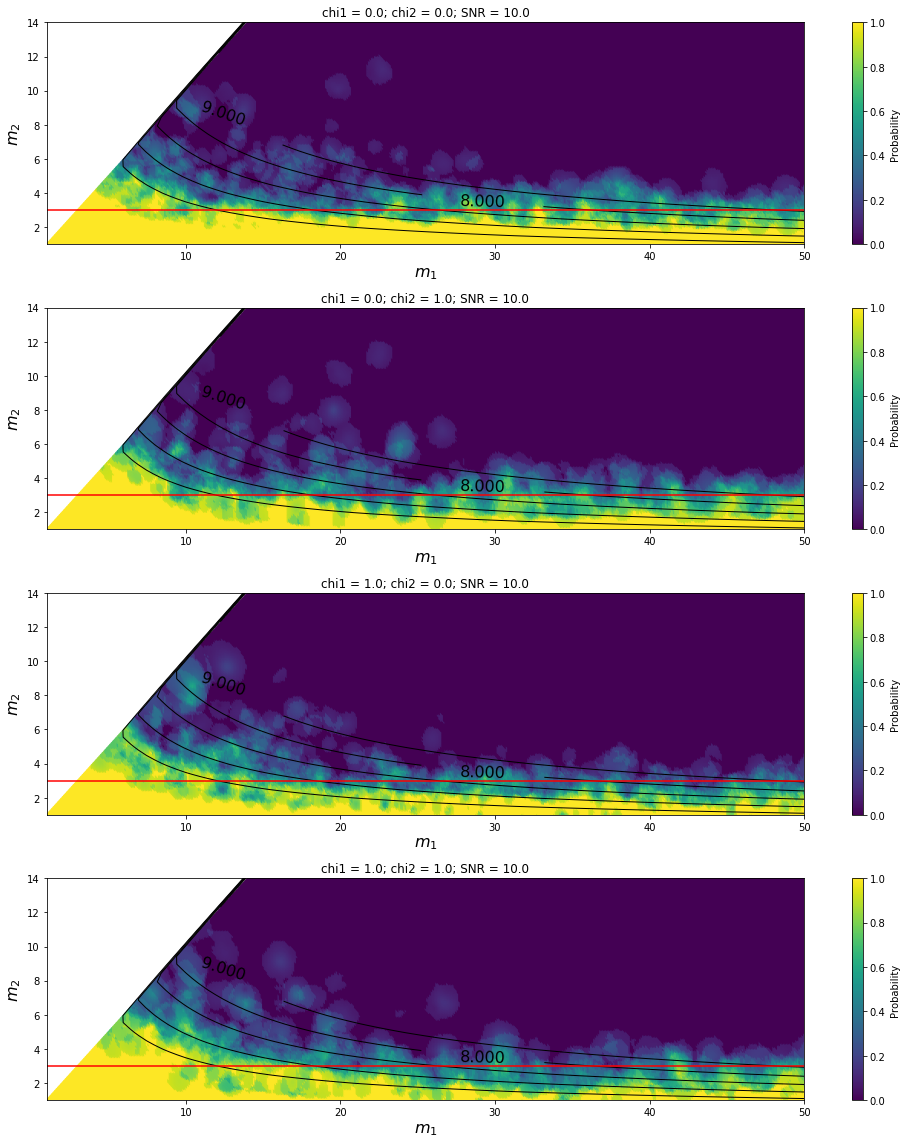
Parameter sweep for the HasNS predictor¶
[5]:
fig = plt.figure(figsize=(14, 16))
for idx, spin_vals in enumerate(spins):
SNR = 10.
title = "chi1 = {0}; chi2 = {1}; SNR = {2}".format(spin_vals['chi1'],
spin_vals['chi2'],
SNR)
SNR *= np.ones(np.sum(mask))
chi1 = spin_vals['chi1'] * np.ones(np.sum(mask))
chi2 = spin_vals['chi2'] * np.ones(np.sum(mask))
# make predictions and make plots
param_sweep_features = np.stack([t['mass1'], t['mass2'], chi1, chi2, SNR]).T
ns_predictions = clf_ns.predict_proba(param_sweep_features).T[1]
make_plots(param_sweep_features, ns_predictions, title, (fig, idx+1))
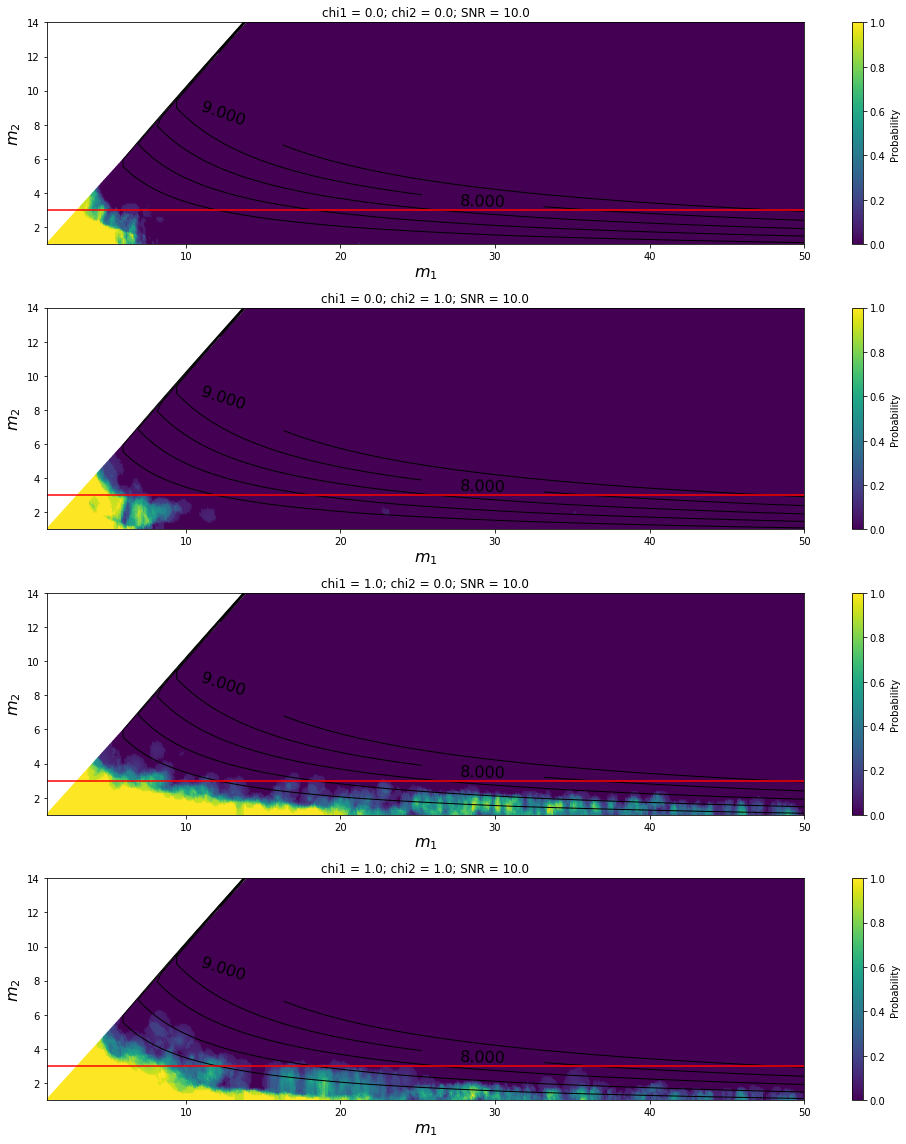
Parameter sweep for the HasRemnant predictor¶
[6]:
fig = plt.figure(figsize=(14, 16))
for idx, spin_vals in enumerate(spins):
SNR = 10.
title = "chi1 = {0}; chi2 = {1}; SNR = {2}".format(spin_vals['chi1'],
spin_vals['chi2'],
SNR)
SNR *= np.ones(np.sum(mask))
chi1 = spin_vals['chi1'] * np.ones(np.sum(mask))
chi2 = spin_vals['chi2'] * np.ones(np.sum(mask))
# make predictions and make plots
param_sweep_features = np.stack([t['mass1'], t['mass2'], chi1, chi2, SNR]).T
remnant_matter_predictions = clf_em.predict_proba(param_sweep_features).T[1]
make_plots(param_sweep_features,
remnant_matter_predictions,
title,
(fig, idx+1))
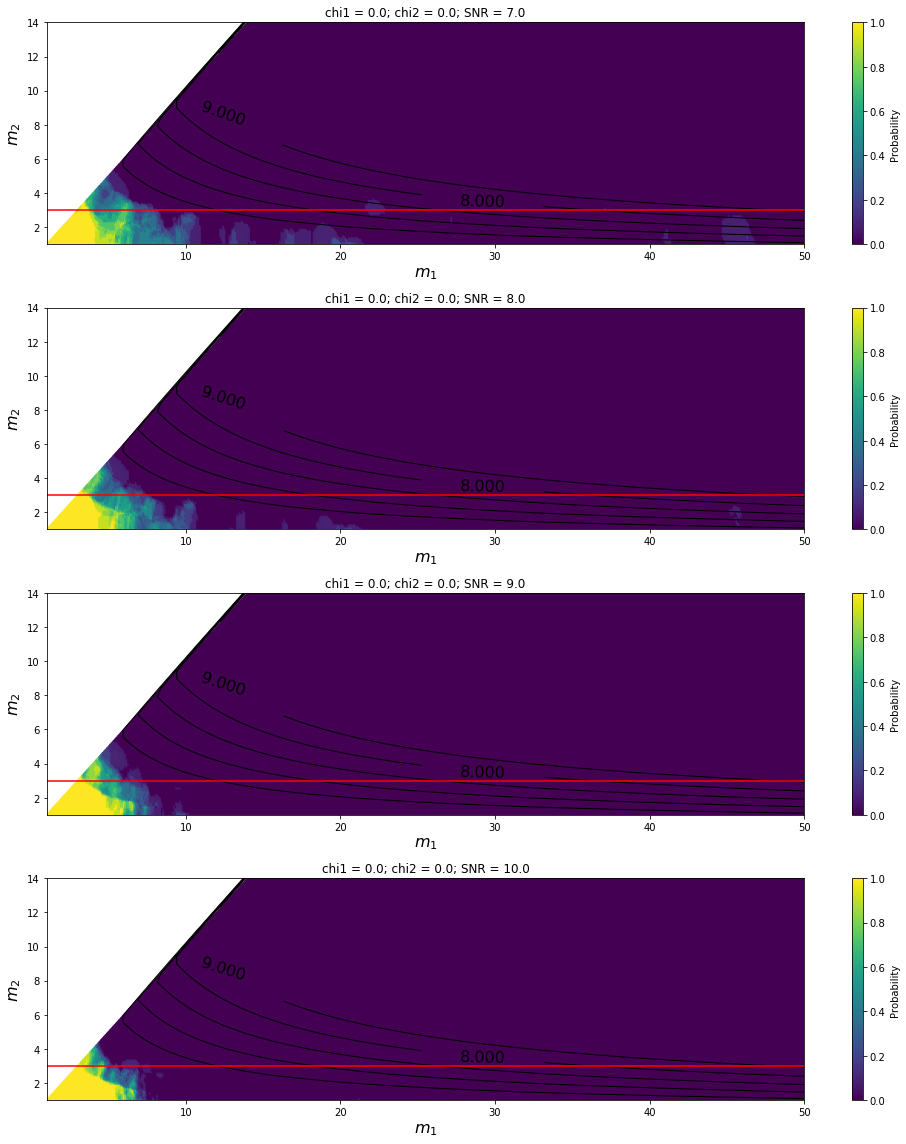
Variation with SNR for HasNS predictor¶
[7]:
fig = plt.figure(figsize=(14, 16))
signal_to_noise = [7.0, 8.0, 9.0, 10.0]
for idx, SNR in enumerate(signal_to_noise):
spin_1 = 0.
spin_2 = 0.
title = "chi1 = {0}; chi2 = {1}; SNR = {2}".format(spin_1,
spin_2,
SNR)
SNR *= np.ones(np.sum(mask))
chi1 = spin_1 * np.ones(np.sum(mask))
chi2 = spin_2 * np.ones(np.sum(mask))
# make predictions and make plots
param_sweep_features = np.stack([t['mass1'], t['mass2'], chi1, chi2, SNR]).T
ns_predictions = clf_ns.predict_proba(param_sweep_features).T[1]
make_plots(param_sweep_features, ns_predictions,
title, (fig, idx+1))
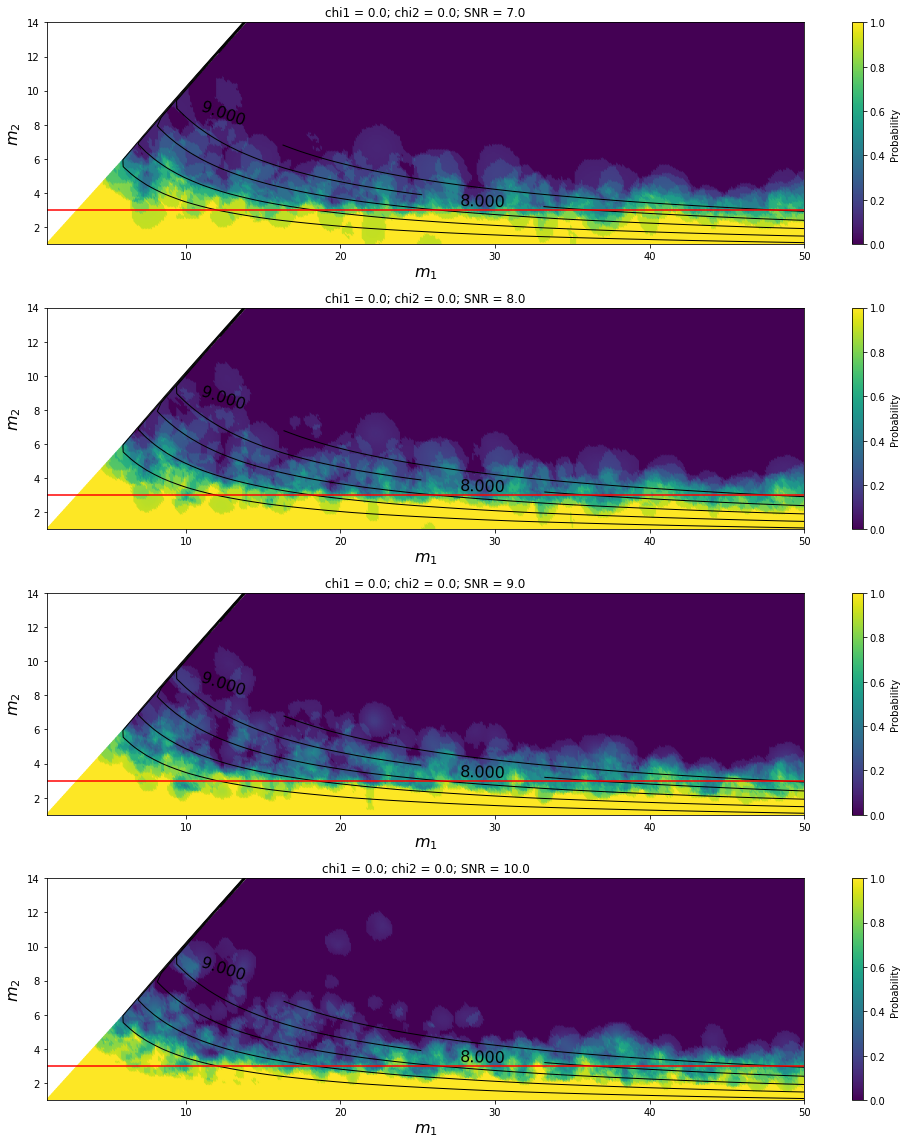
Variation with SNR for HasRemnant predictor¶
[8]:
fig = plt.figure(figsize=(14, 16))
signal_to_noise = [7.0, 8.0, 9.0, 10.0]
for idx, SNR in enumerate(signal_to_noise):
spin_1 = 0.
spin_2 = 0.
title = "chi1 = {0}; chi2 = {1}; SNR = {2}".format(spin_1,
spin_2,
SNR)
SNR *= np.ones(np.sum(mask))
chi1 = spin_1 * np.ones(np.sum(mask))
chi2 = spin_2 * np.ones(np.sum(mask))
# make predictions and make plots
param_sweep_features = np.stack([t['mass1'], t['mass2'], chi1, chi2, SNR]).T
remnant_matter_predictions = clf_em.predict_proba(param_sweep_features).T[1]
make_plots(param_sweep_features,remnant_matter_predictions,
title, (fig, idx+1))