Fitting a model to data with both x and y errors with Bilby
Usually when we fit a model to data with a Gaussian Likelihood we assume that we know x values exactly. This is almost never the case. Here we show how to fit a model with errors in both x and y.
[1]:
import bilby
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
Simulate data
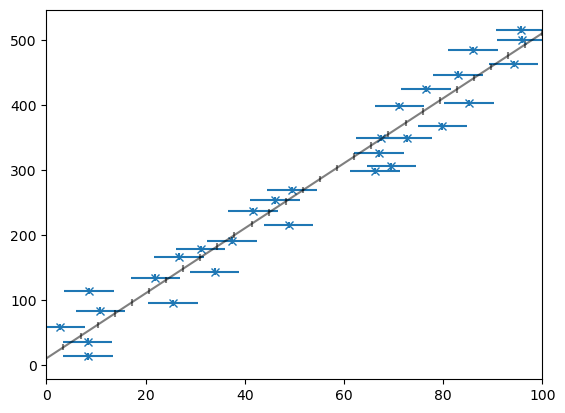
First we create the data and plot it
[2]:
# define our model, a line
def model(x, m, c, **kwargs):
y = m * x + c
return y
# make a function to create and plot our data
def make_data(points, m, c, xerr, yerr, seed):
np.random.seed(int(seed))
xtrue = np.linspace(0, 100, points)
ytrue = model(x=xtrue, m=m, c=c)
xerr_vals = xerr * np.random.randn(points)
yerr_vals = yerr * np.random.randn(points)
xobs = xtrue + xerr_vals
yobs = ytrue + yerr_vals
plt.errorbar(xobs, yobs, xerr=xerr, yerr=yerr, fmt="x")
plt.errorbar(xtrue, ytrue, yerr=yerr, color="black", alpha=0.5)
plt.xlim(0, 100)
plt.show()
plt.close()
data = {
"xtrue": xtrue,
"ytrue": ytrue,
"xobs": xobs,
"yobs": yobs,
"xerr": xerr,
"yerr": yerr,
}
return data
data = make_data(points=30, m=5, c=10, xerr=5, yerr=5, seed=123)
Define our prior and sampler settings
Now lets set up the prior and bilby output directory/sampler settings
[3]:
# setting up bilby priors
priors = dict(
m=bilby.core.prior.Uniform(0, 30, "m"), c=bilby.core.prior.Uniform(0, 30, "c")
)
sampler_kwargs = dict(priors=priors, sampler="bilby_mcmc", nsamples=1000, printdt=5, outdir="outdir", verbose=False, clean=True)
Fit with exactly known x-values
Our first step is to recover the straight line using a simple Gaussian Likelihood that only takes into account the y errors. Under the assumption we know x exactly. In this case, we pass in xtrue for x
[4]:
known_x = bilby.core.likelihood.GaussianLikelihood(
x=data["xtrue"], y=data["yobs"], func=model, sigma=data["yerr"]
)
result_known_x = bilby.run_sampler(
likelihood=known_x,
label="known_x",
**sampler_kwargs,
)
05:18 bilby INFO : Running for label 'known_x', output will be saved to 'outdir'
05:18 bilby INFO : Analysis priors:
05:18 bilby INFO : m=Uniform(minimum=0, maximum=30, name='m', latex_label='m', unit=None, boundary=None)
05:18 bilby INFO : c=Uniform(minimum=0, maximum=30, name='c', latex_label='c', unit=None, boundary=None)
05:18 bilby INFO : Analysis likelihood class: <class 'bilby.core.likelihood.GaussianLikelihood'>
05:18 bilby INFO : Analysis likelihood noise evidence: nan
05:18 bilby INFO : Single likelihood evaluation took 8.403e-05 s
05:18 bilby INFO : Using sampler Bilby_MCMC with kwargs {'nsamples': 1000, 'nensemble': 1, 'pt_ensemble': False, 'ntemps': 1, 'Tmax': None, 'Tmax_from_SNR': 20, 'initial_betas': None, 'adapt': True, 'adapt_t0': 100, 'adapt_nu': 10, 'pt_rejection_sample': False, 'burn_in_nact': 10, 'thin_by_nact': 1, 'fixed_discard': 0, 'autocorr_c': 5, 'L1steps': 100, 'L2steps': 3, 'printdt': 5, 'check_point_delta_t': 1800, 'min_tau': 1, 'proposal_cycle': 'default', 'stop_after_convergence': False, 'fixed_tau': None, 'tau_window': None, 'evidence_method': 'stepping_stone', 'initial_sample_method': 'prior', 'initial_sample_dict': None}
05:18 bilby INFO : Initializing BilbyPTMCMCSampler with:
Convergence settings: ConvergenceInputs(autocorr_c=5, burn_in_nact=10, thin_by_nact=1, fixed_discard=0, target_nsamples=1000, stop_after_convergence=False, L1steps=100, L2steps=3, min_tau=1, fixed_tau=None, tau_window=None)
Parallel-tempering settings: ParallelTemperingInputs(ntemps=1, nensemble=1, Tmax=None, Tmax_from_SNR=20, initial_betas=None, adapt=True, adapt_t0=100, adapt_nu=10, pt_ensemble=False)
proposal_cycle: default
pt_rejection_sample: False
05:18 bilby INFO : Setting parallel tempering inputs=ParallelTemperingInputs(ntemps=1, nensemble=1, Tmax=None, Tmax_from_SNR=20, initial_betas=None, adapt=True, adapt_t0=100, adapt_nu=10, pt_ensemble=False)
05:18 bilby INFO : Initializing BilbyPTMCMCSampler with:ntemps=1, nensemble=1, pt_ensemble=False, initial_betas=[1], initial_sample_method=prior, initial_sample_dict=None
05:18 bilby INFO : Using initial sample {'m': 7.554705855330316, 'c': 4.605823628092564}
05:18 bilby INFO : Using ProposalCycle:
AdaptiveGaussianProposal(acceptance_ratio:nan,n:0,scale:1,)
DifferentialEvolutionProposal(acceptance_ratio:nan,n:0,)
UniformProposal(acceptance_ratio:nan,n:0,)
KDEProposal(acceptance_ratio:nan,n:0,trained:0,)
FisherMatrixProposal(acceptance_ratio:nan,n:0,scale:1,)
GMMProposal(acceptance_ratio:nan,n:0,trained:0,)
05:18 bilby INFO : Setting convergence_inputs=ConvergenceInputs(autocorr_c=5, burn_in_nact=10, thin_by_nact=1, fixed_discard=0, target_nsamples=1000, stop_after_convergence=False, L1steps=100, L2steps=3, min_tau=1, fixed_tau=None, tau_window=None)
05:18 bilby INFO : Drawing 1000 samples
05:18 bilby INFO : Checkpoint every check_point_delta_t=1800s
05:18 bilby INFO : Print update every printdt=5s
05:18 bilby INFO : Reached convergence: exiting sampling
05:18 bilby INFO : Checkpoint start
05:18 bilby INFO : Written checkpoint file outdir/known_x_resume.pickle
05:18 bilby INFO : Zero-temperature proposals:
05:18 bilby INFO : AdaptiveGaussianProposal(acceptance_ratio:0.23,n:2.9e+04,scale:0.00058,)
05:18 bilby INFO : DifferentialEvolutionProposal(acceptance_ratio:0.47,n:3.1e+04,)
05:18 bilby INFO : UniformProposal(acceptance_ratio:1,n:9.6e+02,)
05:18 bilby INFO : KDEProposal(acceptance_ratio:0.00014,n:2.9e+04,trained:0,)
05:18 bilby INFO : FisherMatrixProposal(acceptance_ratio:0.55,n:2.7e+04,scale:1,)
05:18 bilby INFO : GMMProposal(acceptance_ratio:4.3e-05,n:2.3e+04,trained:0,)
05:18 bilby INFO : Current taus={'m': 1, 'c': 1}
05:18 bilby INFO : Creating diagnostic plots
05:18 bilby INFO : Checkpoint finished
05:18 bilby INFO : Sampling time: 0:00:15.012288
05:18 bilby INFO : Summary of results:
nsamples: 1289
ln_noise_evidence: nan
ln_evidence: nan +/- nan
ln_bayes_factor: nan +/- nan
[5]:
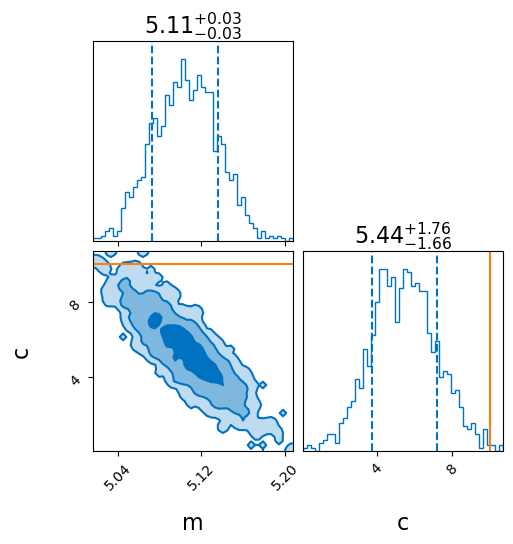
_ = result_known_x.plot_corner(truth=dict(m=5, c=10), titles=True, save=False)
plt.show()
plt.close()
Fit with unmodeled uncertainty in the x-values
As expected this is easy to recover and the sampler does a good job. However this was made too easy - by passing in the ‘true’ values of x. Lets see what happens when we pass in the observed values of x
[6]:
incorrect_x = bilby.core.likelihood.GaussianLikelihood(
x=data["xobs"], y=data["yobs"], func=model, sigma=data["yerr"]
)
result_incorrect_x = bilby.run_sampler(
likelihood=incorrect_x,
label="incorrect_x",
**sampler_kwargs,
)
05:18 bilby INFO : Running for label 'incorrect_x', output will be saved to 'outdir'
05:19 bilby INFO : Analysis priors:
05:19 bilby INFO : m=Uniform(minimum=0, maximum=30, name='m', latex_label='m', unit=None, boundary=None)
05:19 bilby INFO : c=Uniform(minimum=0, maximum=30, name='c', latex_label='c', unit=None, boundary=None)
05:19 bilby INFO : Analysis likelihood class: <class 'bilby.core.likelihood.GaussianLikelihood'>
05:19 bilby INFO : Analysis likelihood noise evidence: nan
05:19 bilby INFO : Single likelihood evaluation took 6.831e-05 s
05:19 bilby INFO : Using sampler Bilby_MCMC with kwargs {'nsamples': 1000, 'nensemble': 1, 'pt_ensemble': False, 'ntemps': 1, 'Tmax': None, 'Tmax_from_SNR': 20, 'initial_betas': None, 'adapt': True, 'adapt_t0': 100, 'adapt_nu': 10, 'pt_rejection_sample': False, 'burn_in_nact': 10, 'thin_by_nact': 1, 'fixed_discard': 0, 'autocorr_c': 5, 'L1steps': 100, 'L2steps': 3, 'printdt': 5, 'check_point_delta_t': 1800, 'min_tau': 1, 'proposal_cycle': 'default', 'stop_after_convergence': False, 'fixed_tau': None, 'tau_window': None, 'evidence_method': 'stepping_stone', 'initial_sample_method': 'prior', 'initial_sample_dict': None}
05:19 bilby INFO : Initializing BilbyPTMCMCSampler with:
Convergence settings: ConvergenceInputs(autocorr_c=5, burn_in_nact=10, thin_by_nact=1, fixed_discard=0, target_nsamples=1000, stop_after_convergence=False, L1steps=100, L2steps=3, min_tau=1, fixed_tau=None, tau_window=None)
Parallel-tempering settings: ParallelTemperingInputs(ntemps=1, nensemble=1, Tmax=None, Tmax_from_SNR=20, initial_betas=None, adapt=True, adapt_t0=100, adapt_nu=10, pt_ensemble=False)
proposal_cycle: default
pt_rejection_sample: False
05:19 bilby INFO : Setting parallel tempering inputs=ParallelTemperingInputs(ntemps=1, nensemble=1, Tmax=None, Tmax_from_SNR=20, initial_betas=None, adapt=True, adapt_t0=100, adapt_nu=10, pt_ensemble=False)
05:19 bilby INFO : Initializing BilbyPTMCMCSampler with:ntemps=1, nensemble=1, pt_ensemble=False, initial_betas=[1], initial_sample_method=prior, initial_sample_dict=None
05:19 bilby INFO : Using initial sample {'m': 12.963788040930169, 'c': 20.214929136508314}
05:19 bilby INFO : Using ProposalCycle:
AdaptiveGaussianProposal(acceptance_ratio:nan,n:0,scale:1,)
DifferentialEvolutionProposal(acceptance_ratio:nan,n:0,)
UniformProposal(acceptance_ratio:nan,n:0,)
KDEProposal(acceptance_ratio:nan,n:0,trained:0,)
FisherMatrixProposal(acceptance_ratio:nan,n:0,scale:1,)
GMMProposal(acceptance_ratio:nan,n:0,trained:0,)
05:19 bilby INFO : Setting convergence_inputs=ConvergenceInputs(autocorr_c=5, burn_in_nact=10, thin_by_nact=1, fixed_discard=0, target_nsamples=1000, stop_after_convergence=False, L1steps=100, L2steps=3, min_tau=1, fixed_tau=None, tau_window=None)
05:19 bilby INFO : Drawing 1000 samples
05:19 bilby INFO : Checkpoint every check_point_delta_t=1800s
05:19 bilby INFO : Print update every printdt=5s
05:19 bilby INFO : Reached convergence: exiting sampling
05:19 bilby INFO : Checkpoint start
05:19 bilby INFO : Written checkpoint file outdir/incorrect_x_resume.pickle
05:19 bilby INFO : Zero-temperature proposals:
05:19 bilby INFO : AdaptiveGaussianProposal(acceptance_ratio:0.23,n:3e+04,scale:0.0045,)
05:19 bilby INFO : DifferentialEvolutionProposal(acceptance_ratio:0.47,n:2.7e+04,)
05:19 bilby INFO : UniformProposal(acceptance_ratio:1,n:9.7e+02,)
05:19 bilby INFO : KDEProposal(acceptance_ratio:7.6e-05,n:2.6e+04,trained:0,)
05:19 bilby INFO : FisherMatrixProposal(acceptance_ratio:0.56,n:2.8e+04,scale:1,)
05:19 bilby INFO : GMMProposal(acceptance_ratio:7.2e-05,n:2.8e+04,trained:0,)
05:19 bilby INFO : Current taus={'m': 1.0, 'c': 1.0}
05:19 bilby INFO : Creating diagnostic plots
05:19 bilby INFO : Checkpoint finished
05:19 bilby INFO : Sampling time: 0:00:15.016030
05:19 bilby INFO : Summary of results:
nsamples: 1299
ln_noise_evidence: nan
ln_evidence: nan +/- nan
ln_bayes_factor: nan +/- nan
[7]:
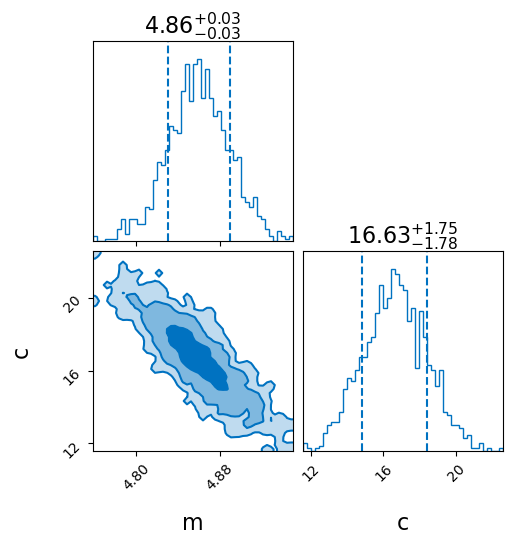
_ = result_incorrect_x.plot_corner(truth=dict(m=5, c=10), titles=True, save=False)
plt.show()
plt.close()
Fit with modeled uncertainty in x-values
This is not good as there is unmodelled uncertainty in our x values. Getting around this requires marginalisation of the true x values or sampling over them. See discussion in section 7 of https://arxiv.org/pdf/1008.4686.pdf.
For this, we will have to define a new likelihood class. By subclassing the base bilby.core.likelihood.Likelihood class we can do this fairly simply.
[8]:
class GaussianLikelihoodUncertainX(bilby.core.likelihood.Likelihood):
def __init__(self, xobs, yobs, xerr, yerr, function):
"""
Parameters
----------
xobs, yobs: array_like
The data to analyse
xerr, yerr: array_like
The standard deviation of the noise
function:
The python function to fit to the data
"""
super(GaussianLikelihoodUncertainX, self).__init__(dict())
self.xobs = xobs
self.yobs = yobs
self.yerr = yerr
self.xerr = xerr
self.function = function
def log_likelihood(self):
variance = (self.xerr * self.parameters["m"]) ** 2 + self.yerr**2
model_y = self.function(self.xobs, **self.parameters)
residual = self.yobs - model_y
ll = -0.5 * np.sum(residual**2 / variance + np.log(variance))
return -0.5 * np.sum(residual**2 / variance + np.log(variance))
[9]:
gaussian_unknown_x = GaussianLikelihoodUncertainX(
xobs=data["xobs"],
yobs=data["yobs"],
xerr=data["xerr"],
yerr=data["yerr"],
function=model,
)
result_unknown_x = bilby.run_sampler(
likelihood=gaussian_unknown_x,
label="unknown_x",
**sampler_kwargs,
)
05:19 bilby INFO : Running for label 'unknown_x', output will be saved to 'outdir'
05:19 bilby INFO : Analysis priors:
05:19 bilby INFO : m=Uniform(minimum=0, maximum=30, name='m', latex_label='m', unit=None, boundary=None)
05:19 bilby INFO : c=Uniform(minimum=0, maximum=30, name='c', latex_label='c', unit=None, boundary=None)
05:19 bilby INFO : Analysis likelihood class: <class '__main__.GaussianLikelihoodUncertainX'>
05:19 bilby INFO : Analysis likelihood noise evidence: nan
05:19 bilby INFO : Single likelihood evaluation took 1.012e-04 s
05:19 bilby INFO : Using sampler Bilby_MCMC with kwargs {'nsamples': 1000, 'nensemble': 1, 'pt_ensemble': False, 'ntemps': 1, 'Tmax': None, 'Tmax_from_SNR': 20, 'initial_betas': None, 'adapt': True, 'adapt_t0': 100, 'adapt_nu': 10, 'pt_rejection_sample': False, 'burn_in_nact': 10, 'thin_by_nact': 1, 'fixed_discard': 0, 'autocorr_c': 5, 'L1steps': 100, 'L2steps': 3, 'printdt': 5, 'check_point_delta_t': 1800, 'min_tau': 1, 'proposal_cycle': 'default', 'stop_after_convergence': False, 'fixed_tau': None, 'tau_window': None, 'evidence_method': 'stepping_stone', 'initial_sample_method': 'prior', 'initial_sample_dict': None}
05:19 bilby INFO : Initializing BilbyPTMCMCSampler with:
Convergence settings: ConvergenceInputs(autocorr_c=5, burn_in_nact=10, thin_by_nact=1, fixed_discard=0, target_nsamples=1000, stop_after_convergence=False, L1steps=100, L2steps=3, min_tau=1, fixed_tau=None, tau_window=None)
Parallel-tempering settings: ParallelTemperingInputs(ntemps=1, nensemble=1, Tmax=None, Tmax_from_SNR=20, initial_betas=None, adapt=True, adapt_t0=100, adapt_nu=10, pt_ensemble=False)
proposal_cycle: default
pt_rejection_sample: False
05:19 bilby INFO : Setting parallel tempering inputs=ParallelTemperingInputs(ntemps=1, nensemble=1, Tmax=None, Tmax_from_SNR=20, initial_betas=None, adapt=True, adapt_t0=100, adapt_nu=10, pt_ensemble=False)
05:19 bilby INFO : Initializing BilbyPTMCMCSampler with:ntemps=1, nensemble=1, pt_ensemble=False, initial_betas=[1], initial_sample_method=prior, initial_sample_dict=None
05:19 bilby INFO : Using initial sample {'m': 23.412862841814835, 'c': 10.922390875963515}
05:19 bilby INFO : Using ProposalCycle:
AdaptiveGaussianProposal(acceptance_ratio:nan,n:0,scale:1,)
DifferentialEvolutionProposal(acceptance_ratio:nan,n:0,)
UniformProposal(acceptance_ratio:nan,n:0,)
KDEProposal(acceptance_ratio:nan,n:0,trained:0,)
FisherMatrixProposal(acceptance_ratio:nan,n:0,scale:1,)
GMMProposal(acceptance_ratio:nan,n:0,trained:0,)
05:19 bilby INFO : Setting convergence_inputs=ConvergenceInputs(autocorr_c=5, burn_in_nact=10, thin_by_nact=1, fixed_discard=0, target_nsamples=1000, stop_after_convergence=False, L1steps=100, L2steps=3, min_tau=1, fixed_tau=None, tau_window=None)
05:19 bilby INFO : Drawing 1000 samples
05:19 bilby INFO : Checkpoint every check_point_delta_t=1800s
05:19 bilby INFO : Print update every printdt=5s
05:19 bilby INFO : Reached convergence: exiting sampling
05:19 bilby INFO : Checkpoint start
05:19 bilby INFO : Written checkpoint file outdir/unknown_x_resume.pickle
05:19 bilby INFO : Zero-temperature proposals:
05:19 bilby INFO : AdaptiveGaussianProposal(acceptance_ratio:0.23,n:3.4e+04,scale:0.0091,)
05:19 bilby INFO : DifferentialEvolutionProposal(acceptance_ratio:0.46,n:3.5e+04,)
05:19 bilby INFO : UniformProposal(acceptance_ratio:1,n:1.2e+03,)
05:19 bilby INFO : KDEProposal(acceptance_ratio:0.00096,n:3.7e+04,trained:0,)
05:19 bilby INFO : FisherMatrixProposal(acceptance_ratio:0.52,n:3.6e+04,scale:1,)
05:19 bilby INFO : GMMProposal(acceptance_ratio:0.0011,n:3.7e+04,trained:0,)
05:19 bilby INFO : Current taus={'m': 1.0, 'c': 1.1}
05:19 bilby INFO : Creating diagnostic plots
05:19 bilby INFO : Checkpoint finished
05:19 bilby INFO : Sampling time: 0:00:20.025516
05:19 bilby INFO : Summary of results:
nsamples: 1690
ln_noise_evidence: nan
ln_evidence: nan +/- nan
ln_bayes_factor: nan +/- nan
[10]:
_ = result_unknown_x.plot_corner(truth=dict(m=5, c=10), titles=True, save=False)
plt.show()
plt.close()
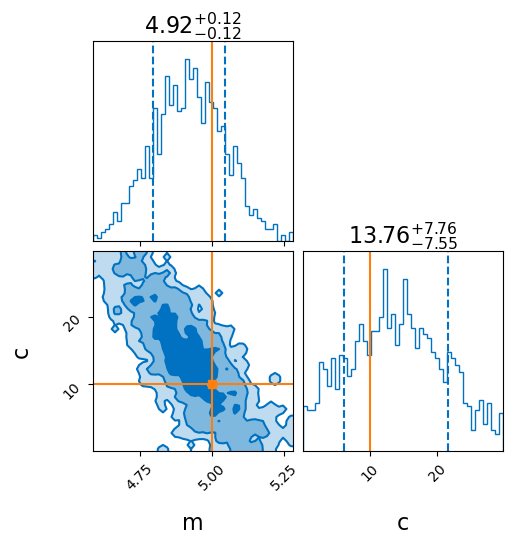
Success! The inferred posterior is consistent with the true values.