[1]:
! rm visualising_the_results/*
rm: cannot remove 'visualising_the_results/*': No such file or directory
Visualising the results
In this tutorial, we demonstrate the plotting tools built-in to bilby and how to extend them. First, we run a simple injection study and return the result object.
[2]:
import bilby
import matplotlib.pyplot as plt
%matplotlib inline
[3]:
duration = 4
sampling_frequency = 2048
outdir = "visualising_the_results"
label = "example"
injection_parameters = dict(
mass_1=36.0,
mass_2=29.0,
a_1=0.4,
a_2=0.3,
tilt_1=0.5,
tilt_2=1.0,
phi_12=1.7,
phi_jl=0.3,
luminosity_distance=1000.0,
theta_jn=0.4,
phase=1.3,
ra=1.375,
dec=-1.2108,
geocent_time=1126259642.413,
psi=2.659,
)
[4]:
# specify waveform arguments
waveform_arguments = dict(
waveform_approximant="IMRPhenomXP", # waveform approximant name
reference_frequency=50.0, # gravitational waveform reference frequency (Hz)
)
# set up the waveform generator
waveform_generator = bilby.gw.waveform_generator.WaveformGenerator(
sampling_frequency=sampling_frequency,
duration=duration,
frequency_domain_source_model=bilby.gw.source.lal_binary_black_hole,
parameters=injection_parameters,
waveform_arguments=waveform_arguments,
)
05:19 bilby INFO : Waveform generator initiated with
frequency_domain_source_model: bilby.gw.source.lal_binary_black_hole
time_domain_source_model: None
parameter_conversion: bilby.gw.conversion.convert_to_lal_binary_black_hole_parameters
[5]:
ifos = bilby.gw.detector.InterferometerList(["H1", "L1"])
ifos.set_strain_data_from_power_spectral_densities(
duration=duration,
sampling_frequency=sampling_frequency,
start_time=injection_parameters["geocent_time"] - 2,
)
_ = ifos.inject_signal(
waveform_generator=waveform_generator, parameters=injection_parameters
)
/opt/conda/envs/python311/lib/python3.11/site-packages/lalsimulation/lalsimulation.py:8: UserWarning: Wswiglal-redir-stdio:
SWIGLAL standard output/error redirection is enabled in IPython.
This may lead to performance penalties. To disable locally, use:
with lal.no_swig_redirect_standard_output_error():
...
To disable globally, use:
lal.swig_redirect_standard_output_error(False)
Note however that this will likely lead to error messages from
LAL functions being either misdirected or lost when called from
Jupyter notebooks.
To suppress this warning, use:
import warnings
warnings.filterwarnings("ignore", "Wswiglal-redir-stdio")
import lal
import lal
05:19 bilby INFO : Injected signal in H1:
05:19 bilby INFO : optimal SNR = 23.78
05:19 bilby INFO : matched filter SNR = 24.32+0.28j
05:19 bilby INFO : mass_1 = 36.0
05:19 bilby INFO : mass_2 = 29.0
05:19 bilby INFO : a_1 = 0.4
05:19 bilby INFO : a_2 = 0.3
05:19 bilby INFO : tilt_1 = 0.5
05:19 bilby INFO : tilt_2 = 1.0
05:19 bilby INFO : phi_12 = 1.7
05:19 bilby INFO : phi_jl = 0.3
05:19 bilby INFO : luminosity_distance = 1000.0
05:19 bilby INFO : theta_jn = 0.4
05:19 bilby INFO : phase = 1.3
05:19 bilby INFO : ra = 1.375
05:19 bilby INFO : dec = -1.2108
05:19 bilby INFO : geocent_time = 1126259642.413
05:19 bilby INFO : psi = 2.659
05:19 bilby INFO : Injected signal in L1:
05:19 bilby INFO : optimal SNR = 19.24
05:19 bilby INFO : matched filter SNR = 19.99+0.16j
05:19 bilby INFO : mass_1 = 36.0
05:19 bilby INFO : mass_2 = 29.0
05:19 bilby INFO : a_1 = 0.4
05:19 bilby INFO : a_2 = 0.3
05:19 bilby INFO : tilt_1 = 0.5
05:19 bilby INFO : tilt_2 = 1.0
05:19 bilby INFO : phi_12 = 1.7
05:19 bilby INFO : phi_jl = 0.3
05:19 bilby INFO : luminosity_distance = 1000.0
05:19 bilby INFO : theta_jn = 0.4
05:19 bilby INFO : phase = 1.3
05:19 bilby INFO : ra = 1.375
05:19 bilby INFO : dec = -1.2108
05:19 bilby INFO : geocent_time = 1126259642.413
05:19 bilby INFO : psi = 2.659
[6]:
# first, set up all priors to be equal to a delta function at their designated value
priors = bilby.gw.prior.BBHPriorDict(injection_parameters.copy())
# then, reset the priors on the masses and luminosity distance to conduct a search over these parameters
priors["mass_1"] = bilby.core.prior.Uniform(25, 40, "mass_1")
priors["mass_2"] = bilby.core.prior.Uniform(25, 40, "mass_2")
priors["luminosity_distance"] = bilby.core.prior.Uniform(
400, 2000, "luminosity_distance"
)
[7]:
# compute the likelihoods
likelihood = bilby.gw.likelihood.GravitationalWaveTransient(
interferometers=ifos, waveform_generator=waveform_generator
)
Set up the sampling
For this case we use the dynesty sampler with 100 live points and the uniform sampling method. While these settings are sufficient for this simple, three-dimensional problem, they often don’t work for more complex cases.
[8]:
result = bilby.core.sampler.run_sampler(
likelihood=likelihood,
priors=priors,
sampler="dynesty",
npoints=100,
injection_parameters=injection_parameters,
outdir=outdir,
label=label,
sample="unif",
)
05:19 bilby INFO : Running for label 'example', output will be saved to 'visualising_the_results'
05:19 bilby INFO : Using lal version 7.6.0
05:19 bilby INFO : Using lal git version Branch: None;Tag: lal-v7.6.0;Id: 43d3b7d8137d11b0dd40905636c9f1628d5cbde2;;Builder: Adam Mercer <adam.mercer@ligo.org>;Repository status: CLEAN: All modifications committed
05:19 bilby INFO : Using lalsimulation version 6.0.0
05:19 bilby INFO : Using lalsimulation git version Branch: None;Tag: lalsimulation-v6.0.0;Id: 5fd08c4d441c0684ae5ab3bffaad442240e30461;;Builder: Adam Mercer <adam.mercer@ligo.org>;Repository status: CLEAN: All modifications committed
05:20 bilby INFO : Analysis priors:
05:20 bilby INFO : mass_1=Uniform(minimum=25, maximum=40, name='mass_1', latex_label='$m_1$', unit=None, boundary=None)
05:20 bilby INFO : mass_2=Uniform(minimum=25, maximum=40, name='mass_2', latex_label='$m_2$', unit=None, boundary=None)
05:20 bilby INFO : luminosity_distance=Uniform(minimum=400, maximum=2000, name='luminosity_distance', latex_label='$d_L$', unit=None, boundary=None)
05:20 bilby INFO : a_1=0.4
05:20 bilby INFO : a_2=0.3
05:20 bilby INFO : tilt_1=0.5
05:20 bilby INFO : tilt_2=1.0
05:20 bilby INFO : phi_12=1.7
05:20 bilby INFO : phi_jl=0.3
05:20 bilby INFO : theta_jn=0.4
05:20 bilby INFO : phase=1.3
05:20 bilby INFO : ra=1.375
05:20 bilby INFO : dec=-1.2108
05:20 bilby INFO : geocent_time=1126259642.413
05:20 bilby INFO : psi=2.659
05:20 bilby INFO : Analysis likelihood class: <class 'bilby.gw.likelihood.base.GravitationalWaveTransient'>
05:20 bilby INFO : Analysis likelihood noise evidence: -8487.918743617563
05:20 bilby INFO : Single likelihood evaluation took 8.795e-03 s
05:20 bilby INFO : Using sampler Dynesty with kwargs {'nlive': 100, 'bound': 'live', 'sample': 'unif', 'periodic': None, 'reflective': None, 'update_interval': 600, 'first_update': None, 'npdim': None, 'rstate': None, 'queue_size': 1, 'pool': None, 'use_pool': None, 'live_points': None, 'logl_args': None, 'logl_kwargs': None, 'ptform_args': None, 'ptform_kwargs': None, 'gradient': None, 'grad_args': None, 'grad_kwargs': None, 'compute_jac': False, 'enlarge': None, 'bootstrap': None, 'walks': 100, 'facc': 0.2, 'slices': None, 'fmove': 0.9, 'max_move': 100, 'update_func': None, 'ncdim': None, 'blob': False, 'save_history': False, 'history_filename': None, 'maxiter': None, 'maxcall': None, 'dlogz': 0.1, 'logl_max': inf, 'n_effective': None, 'add_live': True, 'print_progress': True, 'print_func': <bound method Dynesty._print_func of <bilby.core.sampler.dynesty.Dynesty object at 0x7fe38d0b7890>>, 'save_bounds': False, 'checkpoint_file': None, 'checkpoint_every': 60, 'resume': False, 'seed': None}
05:20 bilby INFO : Checkpoint every check_point_delta_t = 600s
05:20 bilby INFO : Using dynesty version 2.1.4
05:20 bilby INFO : Live-point based bound method requested with dynesty sample 'unif', overwriting to 'multi'
05:20 bilby INFO : Resume file visualising_the_results/example_resume.pickle does not exist.
05:20 bilby INFO : Generating initial points from the prior
05:21 bilby INFO : Written checkpoint file visualising_the_results/example_resume.pickle
05:21 bilby INFO : Rejection sampling nested samples to obtain 341 posterior samples
05:21 bilby INFO : Sampling time: 0:01:45.851704
05:21 bilby INFO : Summary of results:
nsamples: 341
ln_noise_evidence: -8487.919
ln_evidence: -8000.739 +/- 0.296
ln_bayes_factor: 487.180 +/- 0.296
Corner plots
Now lets make some corner plots. You can easily generate a corner plot using result.plot_corner() like this:
[9]:
result.plot_corner(save=False)
plt.show()
plt.close()
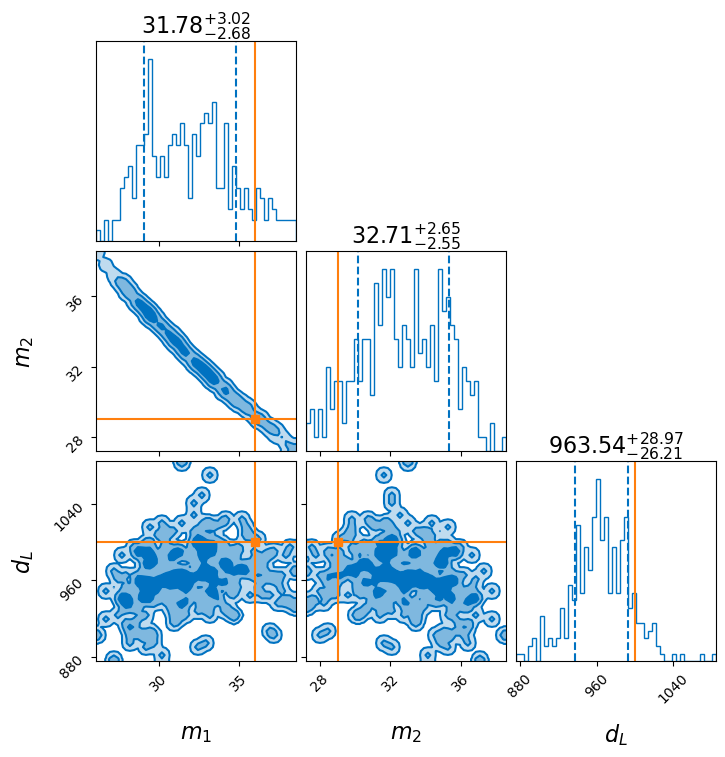
In a notebook, this figure will display. But by default the file is also saved to visualising_the_result/example_corner.png. If you change the label to something more descriptive then the example here will of course be replaced.
Waveform Reconstruction plot
Some plots specific to compact binary coalescence parameter estimation results can be created by re-loading the result as a CBCResult:
[10]:
from bilby.gw.result import CBCResult
cbc_result = CBCResult.from_json("visualising_the_results/example_result.json")
for ifo in ifos:
cbc_result.plot_interferometer_waveform_posterior(
interferometer=ifo, n_samples=500, save=False
)
plt.show()
plt.close()
05:21 bilby INFO : Generating waveform figure for H1
05:21 bilby INFO : Waveform generator initiated with
frequency_domain_source_model: bilby.gw.source.lal_binary_black_hole
time_domain_source_model: None
parameter_conversion: bilby.gw.conversion.convert_to_lal_binary_black_hole_parameters

05:22 bilby INFO : Generating waveform figure for L1
05:22 bilby INFO : Waveform generator initiated with
frequency_domain_source_model: bilby.gw.source.lal_binary_black_hole
time_domain_source_model: None
parameter_conversion: bilby.gw.conversion.convert_to_lal_binary_black_hole_parameters

Marginal Distribution plots
These plots just show the 1D histograms for each parameter
[11]:
result.plot_marginals()
05:22 bilby INFO : Plotting mass_1 marginal distribution
05:22 bilby INFO : Plotting mass_1 marginal distribution
05:22 bilby INFO : Plotting mass_2 marginal distribution
05:22 bilby INFO : Plotting mass_2 marginal distribution
05:22 bilby INFO : Plotting luminosity_distance marginal distribution
05:22 bilby INFO : Plotting luminosity_distance marginal distribution
05:22 bilby INFO : Plotting a_1 marginal distribution
05:22 bilby INFO : Plotting a_1 marginal distribution
05:22 bilby INFO : Plotting a_2 marginal distribution
05:22 bilby INFO : Plotting a_2 marginal distribution
05:22 bilby INFO : Plotting tilt_1 marginal distribution
05:22 bilby INFO : Plotting tilt_1 marginal distribution
05:22 bilby INFO : Plotting tilt_2 marginal distribution
05:22 bilby INFO : Plotting tilt_2 marginal distribution
05:22 bilby INFO : Plotting phi_12 marginal distribution
05:22 bilby INFO : Plotting phi_12 marginal distribution
05:22 bilby INFO : Plotting phi_jl marginal distribution
05:22 bilby INFO : Plotting phi_jl marginal distribution
05:22 bilby INFO : Plotting theta_jn marginal distribution
05:22 bilby INFO : Plotting theta_jn marginal distribution
05:22 bilby INFO : Plotting phase marginal distribution
05:22 bilby INFO : Plotting phase marginal distribution
05:22 bilby INFO : Plotting ra marginal distribution
05:22 bilby INFO : Plotting ra marginal distribution
05:22 bilby INFO : Plotting dec marginal distribution
05:22 bilby INFO : Plotting dec marginal distribution
05:22 bilby INFO : Plotting geocent_time marginal distribution
05:22 bilby INFO : Plotting geocent_time marginal distribution
05:22 bilby INFO : Plotting psi marginal distribution
05:22 bilby INFO : Plotting psi marginal distribution
05:22 bilby INFO : Plotting log_likelihood marginal distribution
05:22 bilby INFO : Plotting log_likelihood marginal distribution
05:22 bilby INFO : Plotting log_prior marginal distribution
05:22 bilby INFO : Plotting log_prior marginal distribution
Customizing corner plots
You may also want to plot a subset of the parameters, or perhaps add the injection_paramters as lines to check if you recovered them correctly. All this can be done through plot_corner. Under the hood, plot_corner uses corner, and all the keyword arguments passed to plot_corner are passed through.
Adding injection parameters to the plot
In the previous plot, you’ll notice bilby added the injection parameters to the plot by default. You can switch this off by setting truth=None when you call plot_corner. Or to add different injection parameters to the plot, just pass this as a keyword argument for truth. In this example, we just add a line for the luminosity distance by passing a dictionary of the value we want to display.
[12]:
result.plot_corner(truth=dict(luminosity_distance=201))
plt.show()
plt.close()
Plot a subset of the corner plot
Or, to plot just a subset of parameters, just pass a list of the names you want.
[13]:
result.plot_corner(
parameters=["mass_1", "mass_2"], filename="{}/subset.png".format(outdir)
)
plt.show()
plt.close()
Notice here, we also passed in a keyword argument filename=, this overwrites the default filename and instead saves the file as visualising_the_results/subset.png. Useful if you want to create lots of different plots.
Alternative
If you would prefer to do the plotting yourself, you can get hold of the samples and the ordering as follows and then plot with a different module. Here is an example using the corner package
[14]:
import corner
samples = result.samples
labels = result.parameter_labels
fig = corner.corner(samples, labels=labels)
plt.show()
plt.close()
